
Hi, I'm Nataly Smith
I specialize in full-stack app development, mathematical modeling, with expertise in cloud technologies/infrastructure and pipeline architecture.
About & Expertise
Senior Software Developer who specializes in full-stack vertical web development, mathematical modeling, with expertise in cloud technologies/infrastructure and pipeline architecture.
I blend cutting-edge technology with mathematical rigor to solve complex problems across multiple domains. From implementing DeepMind's AlphaTensor algorithm in LAPACK to building privacy-first genomic platforms, my work spans the intersection of AI, scientific computing, and real-world applications.
My expertise extends from low-level C and C++ optimization to high-level Python development, modern C# applications, Rust/Tauri desktop applications, Flutter mobile development, and AI-powered full-stack systems.
Full-Stack Vertical Development
Building complete vertical solutions across web, mobile, and desktop platforms.
Cloud Infrastructure
Multi-cloud implementation with Azure, AWS, and Google Cloud Platform.
Mathematical Modeling
Computational modeling and machine learning for research and industry.
AI & Scientific Computing
Advanced algorithms from research to production systems.
Technical Skills
My core expertise spans multiple languages, frameworks, and platforms. I primarily use Python, C#, and custom Bash/Shell scripting, with experience across many other languages and cutting-edge frameworks for professional development.
Languages
- Python
- R
- C#
- C
- C++
- TypeScript
- JavaScript
- Dart
- Fortran
- GDScript
- VB
- Bash
- Matlab
- HTML
- CSS
- SQL
- Racket
- Lisp
Frameworks & Libraries
- React
- Next.js
- Tauri
- Flutter
- Electron
- .NET
- WPF/XAML
- Scikit
- OpenCV
- Flask
- LangGraph
- NextFlow
- Luigi
Tools & Platforms
- Docker
- Git
- Firebase
- Godot
- PostgreSQL
- IIS
- Visual Studio
- VS Code
- Vim
- Linux
- Google Cloud Platform
- Azure
- AWS
- JIRA
- GitHub
- Bitbucket
AI & Scientific Computing
- Machine Learning
- AI/ML
- LangGraph
- GPU Optimization
- Scientific Computing
- Linear Algebra
- Performance Optimization
- Mathematical Modeling
- Computational Biology
Application Development
- Full-Stack Development
- Mobile App Development
- Desktop Applications
- Game Development
- Cross-Platform
- Real-time Systems
- Privacy-First Design
- Genomics Applications
Software Engineering
- OOP
- SOLID
- Agile Development
- Unit Testing
- Integration Testing
- Real-time Collaboration
- Pipeline Architecture
- Cloud Infrastructure
- Multiplayer Systems
Relevant Experience
My career path and key milestones that have shaped my expertise and approach to software development.
Senior Software Developer, AI Platform
Function Health
Austin, TX
Developing and integrating LLMs and agentic workflows into modern applications.
- Optimization of LLM-based agentic workflows for improved productivity and efficiency.
- Development of customer-facing applications that leverage LLMs and agentic workflows.
- Specializing in advanced AI-first development of enterprise grade applications.
Fellowship: Cohort II AI Challenger
Gauntlet AI
Austin, TX
Senior software engineer and second cohort member in highly selective AI training fellowship program.
- Senior software engineer and second cohort member in highly selective AI training fellowship program
- Developing and integrating LLMs and agentic workflows into modern applications
- Collaborating with top-tier engineers on innovative AI-first projects and AI-assisted research
- Specializing in advanced AI-first development of enterprise grade applications
Senior Software Developer
Bruker Cellular Analysis
Remote
Leading development of cloud-native applications and infrastructure automation tools.
- Completed full-scale Azure DevOps migration in just 3 months by coordinating team efforts and creating custom tools
- Re-architectured long-running critical application, improving performance by 400% through code optimization
- Decreased data processing algorithm runtime by 75% with code rewrite, meeting critical product specifications
- Built required testing functionality ahead of schedule, expediting product release by 2 weeks
Software Developer
Bruker Cellular Analysis
Remote
Main developer in creating research tools for next-gen sequencing and genomic analysis.
- Main developer in creating research tools for next-gen sequencing and genomic analysis
- Independently developed a web portal that enabled interdepartmental teams to access research tools
- Introduced automated unit testing and integration testing into CICD pipelines
- Incorporated C++ computer vision libraries to improve sequencing technologies
Computer Systems Engineer
McMaster-Carr
Elmhurst, IL
Full-stack development and system integration for business-critical applications.
- Improved company knowledge sharing and education by creating an internal communications hub
- Delivered business value by developing website features end to end, including Database Schemas and APIs
- Integrated legacy systems to interface with modern business-critical applications
- Worked with legacy SDLC, testing, and release schedules and architecture
Directed Research: Miller-Jensen Laboratory
Yale University
New Haven, CT
Computational research focused on HIV gene expression modeling and bioinformatics analysis.
- Built predictive computational models to discover activation behavior of latent HIV using NFsim
- Generated parameter-phenotype interactome network based on mRNA and protein expression levels
- Utilized R and MATLAB programming to analyze HIV expression data and visualize trends
- Published research findings in peer-reviewed computational biology journal
Bachelor of Science - Biomedical Engineering
Yale University
New Haven, CT
Focus on Biocomputation with research experience in computational modeling and bioinformatics.
- GPA: 3.64 with focus on Biocomputation
- Cum Laude Graduate, National Hispanic Scholar, Vincent Cordova Diversity Award
- Yale CBIT Healthcare Hackathon Runner-Up, Tsai City Accelerator program participant
- Fluent in English, Spanish, French
Senior Software Developer, AI Platform
Function Health
Austin, TX
Developing and integrating LLMs and agentic workflows into modern applications.
- Optimization of LLM-based agentic workflows for improved productivity and efficiency.
- Development of customer-facing applications that leverage LLMs and agentic workflows.
- Specializing in advanced AI-first development of enterprise grade applications.
Fellowship: Cohort II AI Challenger
Gauntlet AI
Austin, TX
Senior software engineer and second cohort member in highly selective AI training fellowship program.
- Senior software engineer and second cohort member in highly selective AI training fellowship program
- Developing and integrating LLMs and agentic workflows into modern applications
- Collaborating with top-tier engineers on innovative AI-first projects and AI-assisted research
- Specializing in advanced AI-first development of enterprise grade applications
Senior Software Developer
Bruker Cellular Analysis
Remote
Leading development of cloud-native applications and infrastructure automation tools.
- Completed full-scale Azure DevOps migration in just 3 months by coordinating team efforts and creating custom tools
- Re-architectured long-running critical application, improving performance by 400% through code optimization
- Decreased data processing algorithm runtime by 75% with code rewrite, meeting critical product specifications
- Built required testing functionality ahead of schedule, expediting product release by 2 weeks
Software Developer
Bruker Cellular Analysis
Remote
Main developer in creating research tools for next-gen sequencing and genomic analysis.
- Main developer in creating research tools for next-gen sequencing and genomic analysis
- Independently developed a web portal that enabled interdepartmental teams to access research tools
- Introduced automated unit testing and integration testing into CICD pipelines
- Incorporated C++ computer vision libraries to improve sequencing technologies
Computer Systems Engineer
McMaster-Carr
Elmhurst, IL
Full-stack development and system integration for business-critical applications.
- Improved company knowledge sharing and education by creating an internal communications hub
- Delivered business value by developing website features end to end, including Database Schemas and APIs
- Integrated legacy systems to interface with modern business-critical applications
- Worked with legacy SDLC, testing, and release schedules and architecture
Directed Research: Miller-Jensen Laboratory
Yale University
New Haven, CT
Computational research focused on HIV gene expression modeling and bioinformatics analysis.
- Built predictive computational models to discover activation behavior of latent HIV using NFsim
- Generated parameter-phenotype interactome network based on mRNA and protein expression levels
- Utilized R and MATLAB programming to analyze HIV expression data and visualize trends
- Published research findings in peer-reviewed computational biology journal
Bachelor of Science - Biomedical Engineering
Yale University
New Haven, CT
Focus on Biocomputation with research experience in computational modeling and bioinformatics.
- GPA: 3.64 with focus on Biocomputation
- Cum Laude Graduate, National Hispanic Scholar, Vincent Cordova Diversity Award
- Yale CBIT Healthcare Hackathon Runner-Up, Tsai City Accelerator program participant
- Fluent in English, Spanish, French
Featured Projects
A selection of projects that demonstrate my technical capabilities across AI-enhanced vertical full-stack app development, mobile development, game development, mathematical modeling, and more.
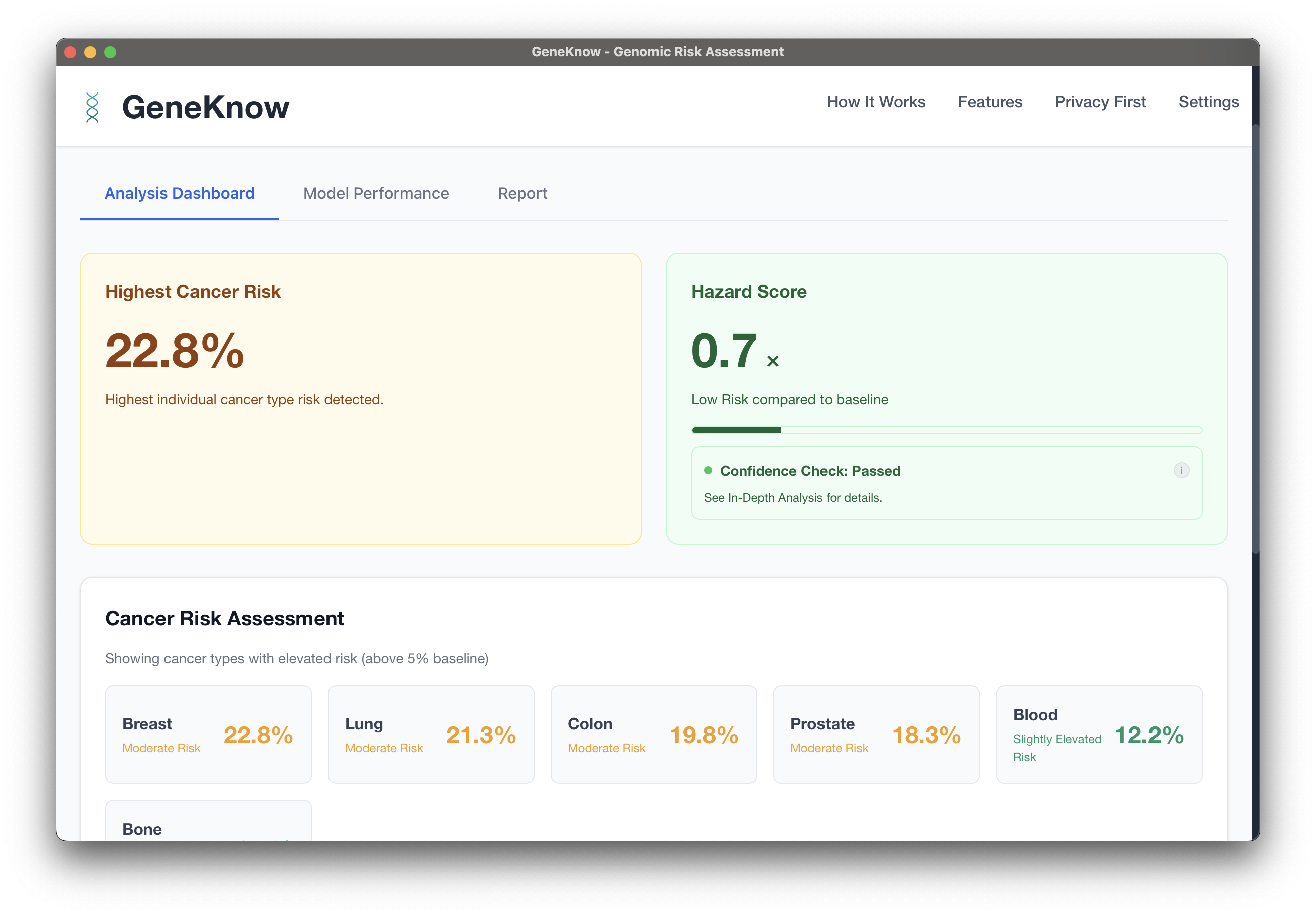
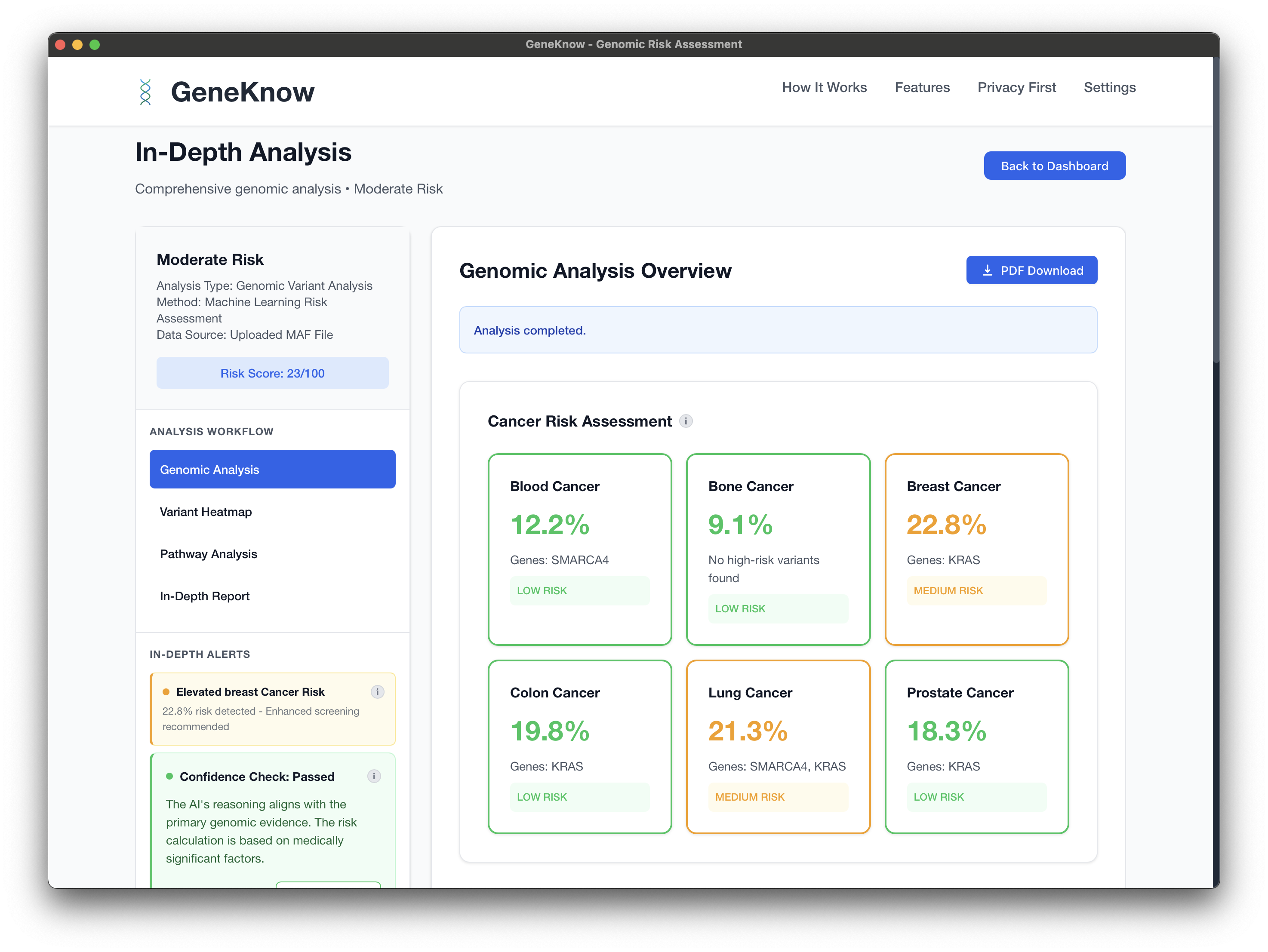
GeneKnow
GeneKnow is a privacy-first and local-first genomic risk assessment platform that processes genetic data entirely on your local machine. No data ever leaves your device, ensuring complete privacy for sensitive genetic information.
LAPACK AI
The first complete, open-source implementation of DeepMind's AlphaTensor 4x4 matrix multiplication algorithm in LAPACK. This landmark computational mathematics project implements a 49-operation algorithm with professional-grade precision, featuring multi-phase CPU optimizations, OpenCL GPU acceleration and kernel implementation on NVIDIA hardware, and seamless LAPACK integration.
MarketSnap
MarketSnap enables farmers-market vendors to share real-time 'fresh-stock' photos and 5-second clips that work offline first, sync transparently when connectivity returns, and auto-expire after 24 hours—driving foot traffic before produce spoils. A cross-platform mobile application built with Flutter and Firebase backend, supporting both iOS and Android platforms.
Children of Singularity
A 2.5D/3D multiplayer sci-fi salvage simulation inspired by Moebius, Planetes, and Nausicaä. Players explore cluttered orbital zones, collect and trade space debris, upgrade their ships (or themselves), and gradually uncover an unsettling AI-controlled ecosystem.
Personyx
Personyx is a compliant desktop app that ingests raw customer-interview transcripts, clusters insights by persona, scores new PRDs for evidence, and lets devs/PMs chat with persona bots while they work. Give makers instant, persona-specific proof that a feature is worth building—before they write code—and live feedback while they do.
FunnelFluent
FunnelFluent AI: Grammarly for sales funnels. Grammarly-like application made for professional making business proposals or marketing materials. A Next.js-based writing assistant powered by Firebase and AI, featuring real-time collaboration, grammar checking, and intelligent writing suggestions.
Additional Projects
Research and engineering projects spanning computational biology, medical devices, and aerospace systems.
Stochastic Computational Modeling of HIV
A transcriptional cycling model that recapitulates chromatin-dependent features of noisy inducible transcription. Published research on computational modeling of variable activation of quiescent HIV infections in T cells.
Simplifying Fracture Treatment: Medical Device Set
Developed a medical device that externally fastens Kirschner wires to prevent complications in fracture malformation and sequestering within the body. Prevents wire migration away from fracture sites.
Project Rocket: Yale Undergraduate Aerospace
Designed and constructed hybrid solid-liquid fuel rockets for national IREC competition. Engineering and fabrication of YUAA's rocket with focus on propulsion systems and aerodynamics.
Bioinformatics of Protein-Protein Interactions
Independent summer computational research visualizing protein-protein interactions using multiple bioinformatic tools. Applied linear regression and random models to TCGA High-Throughput Human data for gene enrichment analyses.
Get in Touch
Interested in working together? Reach out to discuss potential projects or opportunities.
Let's Connect
Ready to discuss your next project? I'd love to hear from you.
Click the button above to open your email client, or reach out to me directly at nmmsoftware@gmail.com